Making poorly described data FAIR-er using GenAI
This small experiment looks at whether Generative AI tools, in this case ChatGPT 4 can improve the metadata (and thus FAIR-ness) of poorly described published datasets.

Generalist data publishing repositories such as Figshare or Zenodo often have little curation. As a result, we see lots of data made available that is potentially very valuable to the community, but not very well described. This small experiment looks at whether Generative AI tools, in this case ChatGPT 4 can improve the metadata (and thus FAIR-ness) of poorly described published datasets.
I decided to look at examples where the dataset has been reuse, even without good metadata, suggesting that the paper associated with the data, describes it well. This dataset is a self-published dataset on Figshare. It does not have a descriptive title and has very light metadata
Delgado-Baquerizo, Manuel (2022). ITS. figshare. Dataset. https://doi.org/10.6084/m9.figshare.5923876.v1
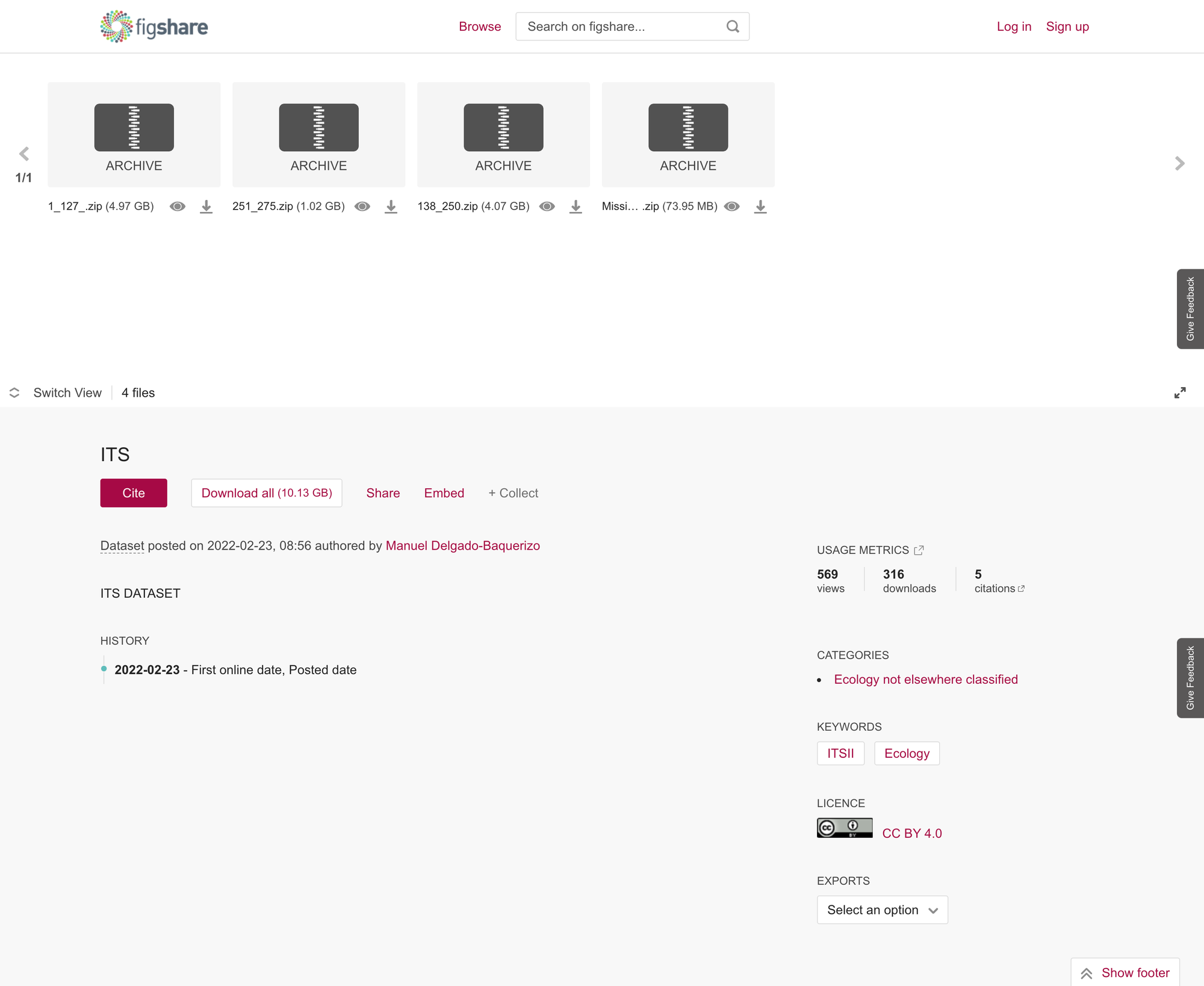
Despite poor metadata, the dataset itself has been cited several times. Using Dimensions.ai we can see that of the 4 papers citing the dataset, 3 have the same author and one is a completely different set of authors. We can assume that the earliest paper to cite the dataset is the one that describes it.
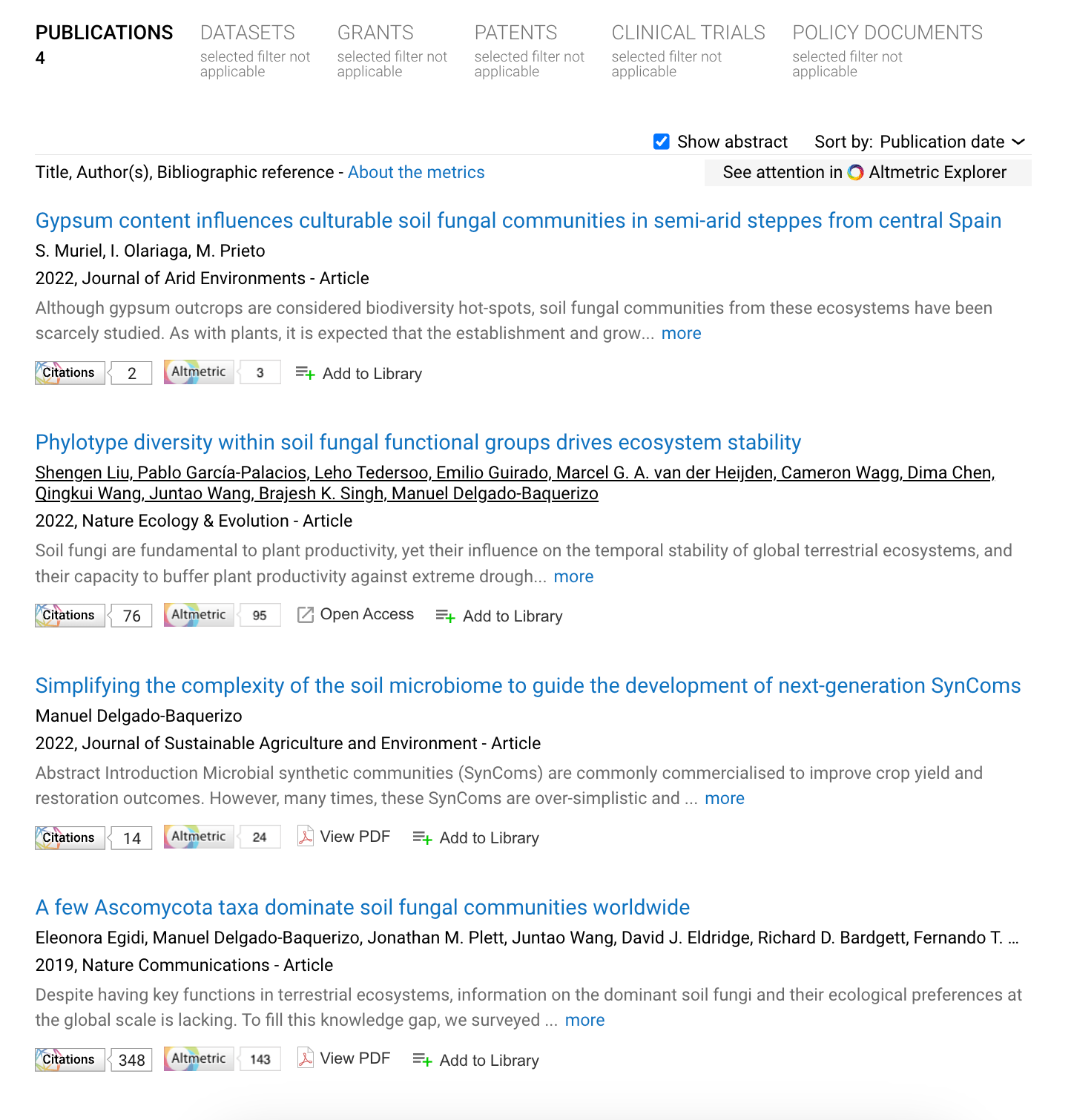
Using a link to the Figshare landing page alone
ChatGPT4
Prompt: Can you create a title, a description, categories and tags to be used in academic literature, for this dataset: https://figshare.com/articles/dataset/ITS/5923876
Response:
- Title: "Comprehensive ITS Dataset for Unexplored Ecological Niches"
- Description: This dataset comprises extensive ITS region sequences, aimed at shedding light on the microbial biodiversity within under-researched ecological niches. Compiled by Manuel Delgado-Baquerizo, it provides a valuable resource for ecological research, especially in areas lacking classification within existing categories. Its extensive coverage and open license make it an essential asset for researchers seeking to understand ecological dynamics more deeply.
- Categories: Ecology, Biodiversity, Microbial Genomics, Environmental Science
- Tags: ITSII, microbial diversity, unclassified ecology, environmental genomics, dataset, open data
Using one of the datasets alone
ChatGPT4
Prompt:
Response:
- Title: "High-Throughput Sequencing Data for Environmental Microbial Analysis"
- Description: This dataset comprises paired-end raw sequencing reads from high-throughput sequencing experiments, encapsulated in six files. The data represents three distinct samples (70, 73, and 123), with each sample's forward (R1) and reverse (R2) reads provided. These reads are essential for comprehensive microbial community analysis, offering insights into the genetic diversity and functional capabilities of environmental samples. The dataset serves as a foundational resource for genomic studies aimed at understanding microbial ecology and evolution.
- Categories: Microbial Genomics, Environmental Genomics, High-Throughput Sequencing, Bioinformatics
- Tags: sequencing data, environmental microbiology, paired-end reads, genomic diversity, FASTQ
Using a link to the Figshare landing page, one of the datasets and the link to the paper
ChatGPT4
Prompt: Can you create a title, a description, categories and tags to be used in academic literature, for this dataset: https://figshare.com/articles/dataset/ITS/5923876 that is referenced in this paper: https://www.nature.com/articles/s41467-019-10373-z
One of the files from the dataset is attached
Response:
- Title: "Global Soil Fungal Dominance: An ITS Dataset Analysis from Diverse Ecosystems"
- Description: This dataset, crucially referenced in the groundbreaking study on global soil fungal communities, contains ITS region sequences that have revealed a few Ascomycota taxa's dominance across various ecosystems worldwide. The research underscores the ecological significance of these taxa in terrestrial biomes, highlighting their roles in nutrient cycling, ecosystem functioning, and potential resilience to environmental changes.
- Categories: Soil Science, Fungal Ecology, Biodiversity, Microbial Ecology
- Tags: Global fungal distribution, Ascomycota, ITS sequencing, soil microbiology, ecological dominance
Conclusion
Each set of metadata is an improvement on the FAIR-ness of the metadata for the Figshare output. Obviously as an n of 1, we cannot read to much into this. But as we scratch the surface of what Generative AI can do for improving metadata in generalist repositories, this result suggest that this is an avenue worth exploring.